Abstract
Background: In the bone marrow (BM) microenvironment, there is a myeloma cell fraction that has acquired chemo-resistance via epigenetic gene expression change. Hypoxic stress is an important factor constituting the BM microenvironment. Recently, we demonstrated that hypoxia-induced microRNA-210 suppressed a ribosomal RNA demethylase, leading to downregulation of IRF4, which is an essential factor in myeloma oncogenesis (Ikeda et al., Cancer Sci 2017). However, myeloma cells showed an anti-apoptotic phenotype without IRF4 under hypoxia, suggesting that there might be another anti-apoptosis factors.
Methods: To determine new anti-apoptosis factor(s) under hypoxia, we conducted gene expression analysis on multiple myeloma (MM) samples (n=4) and cell lines (RPMI-8226, KMS-11, KMS-12-BM, MM.1S, and U266) that cultured under hypoxia (1% O2) for 48 hours. Then, we identify hypoxia-induced gene(s) and conducted functional analysis of the gene(s).
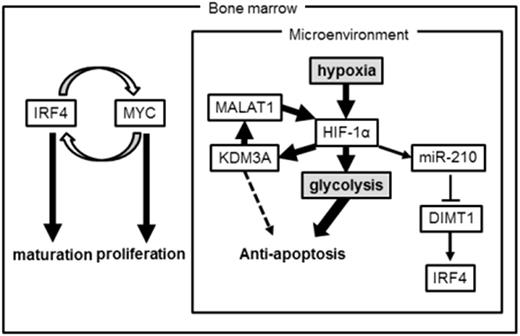
Results: We found that significantly upregulation of glycolytic genes and histone demethylases. Among these, upregulation of H3K9 demethylases, KDM3A and KDM7A were found in all examined cells. We examined a western blot analysis for KDM3A and KDM7A in normoxia- and hypoxia-subjected myeloma cell lines and found that KDM3A showed a stronger increase in hypoxia than that of KDM7A. This result suggested that KDM3A has more potent function in chronic hypoxia than KDM7A does. We confirmed significant upregulation of KDM3A in 15 MM samples cultured in hypoxia (1% O2) for 48 hours by q-RT-PCR. Expression of KDM3A was HIF-1α dependent but not HIF-2α. We found that expression of KDM3A and HIF1A showed a positive correlation in myeloma patient samples. Apoptosis was increased in KDM3A-knockdowned myeloma cells under chronic hypoxia (1% O2) for 72-96 hours. However the target of KDM3A is reported to be IRF4, KDM3A accumulates while IRF4 is suppressed under hypoxia. Therefore we examined the new target gene of KDM3A under hypoxia. We found that the most candidate target of KDM3A under hypoxia was MALAT1. This long non-coding RNA (lncRNA) induces the expression of glycolytic genes such as SLC2A1, PFKFB3, and PFKFB4 via accumulation of HIF-1α. Although IRF4 is crucial factor for myeloma survival under normoxia, it may be switched by HIF-1α-KDM3A-MALAT1 loop under hypoxia (Figure). Activation of this loop contribute to acquiring anti-apoptotic phenotype under hypoxia via upregulation of glycolytic genes.
Conclusion: We conclude KDM3A is promising as a therapeutic target molecule for myeloma cells adapted to the BM microenvironment. Development of KDM3A inhibitor is desired.
Ikeda: TaNeDS (Daiichi Sankyo): Research Funding; Kyowa Kirin: Research Funding; Fujimoto: Research Funding; Novartis: Research Funding; Pfizer: Research Funding; Otsuka: Research Funding; Toyama Kagaku: Research Funding; Chugai: Research Funding; Asahi Kasei: Research Funding; Eisai: Research Funding. Kitadate: Fujimoto: Research Funding; Kyowa Kirin: Research Funding; Eisai: Research Funding; Pfizer: Research Funding; Toyama kagaku: Research Funding; Otsuka: Research Funding; Chugai: Research Funding; Asahi Kasei: Research Funding; Novartis: Research Funding. Abe: Kyowa Kirin: Research Funding; Fujimoto: Research Funding; Novartis: Research Funding; Pfizer: Research Funding; Otsuka: Research Funding; Toyama Kagaku: Research Funding; Chugai: Research Funding; Asahi Kasei: Research Funding; Eisai: Research Funding. Tagawa: TaNeDS (Daiichi Sankyo): Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal